Marvelous Misadventures in Bioinformatics
A blog on some snippets of my work in bioinformatics. Hopefully you find something useful here and avoid stupid mistakes I made.
Statistic annotations with python
“Look what they need to mimic a fraction of our power.”
When we plot graphs comparing quantitative metrics between groups, we often need to add statistical annotations to the plot to denote differences between groups. While programs such as GraphPad Prism and SPSS has built-in tools to add statistical annotations, they lack flexibility and customisability.
This post will introduce programmatic statistical annotations using Python via the statannotations package, using the iris dataset
Prerequisite
- Python >= 3.8
- matplotlib
- seaborn
- statannotations
Installation
pip install statannotations
OR
conda install -c conda-forge statannotations
Usage
- Import packages and load data
import matplotlib.pyplot as plt import seaborn as sns from statannotations.Annotator import Annotator #this is statannotations from itertools import pairwise, permutations, combinations df = sns.load_dataset("iris") - Set the x and y variables, order and combinations of comparisons
x = "species" y = "petal_width" order = ['setosa', 'versicolor', 'virginica'] combo = [(x, y) for x, y in combinations(order, 2)]In this case we want to do a pair-wise comparison between
setosa,versicolorandvirginica - Initialise the plot
ax = sns.boxplot(data=df, x=x, y=y, hue=x, order=order, hue_order=order) -
Initalise the annotator object
The annotator takes your plot object, comparison comaprisons, dataset, x and y variables, and order
annot = Annotator(ax, combo, data=df, x=x, y=y, order=order) -
Configure and apply the test statistic
annot.configure(test='Mann-Whitney', comparisons_correction="Bonferroni", text_format='star', loc='outside', verbose=2) annot.apply_test() -
Annotate the plot
ax, test_results = annot.annotate() plt.show()The
locparameter controls how the annotations are placed,outsidemeans they are placed outside the plot, whileinsidemeans they are placed inside the plotOutside (
loc='outside')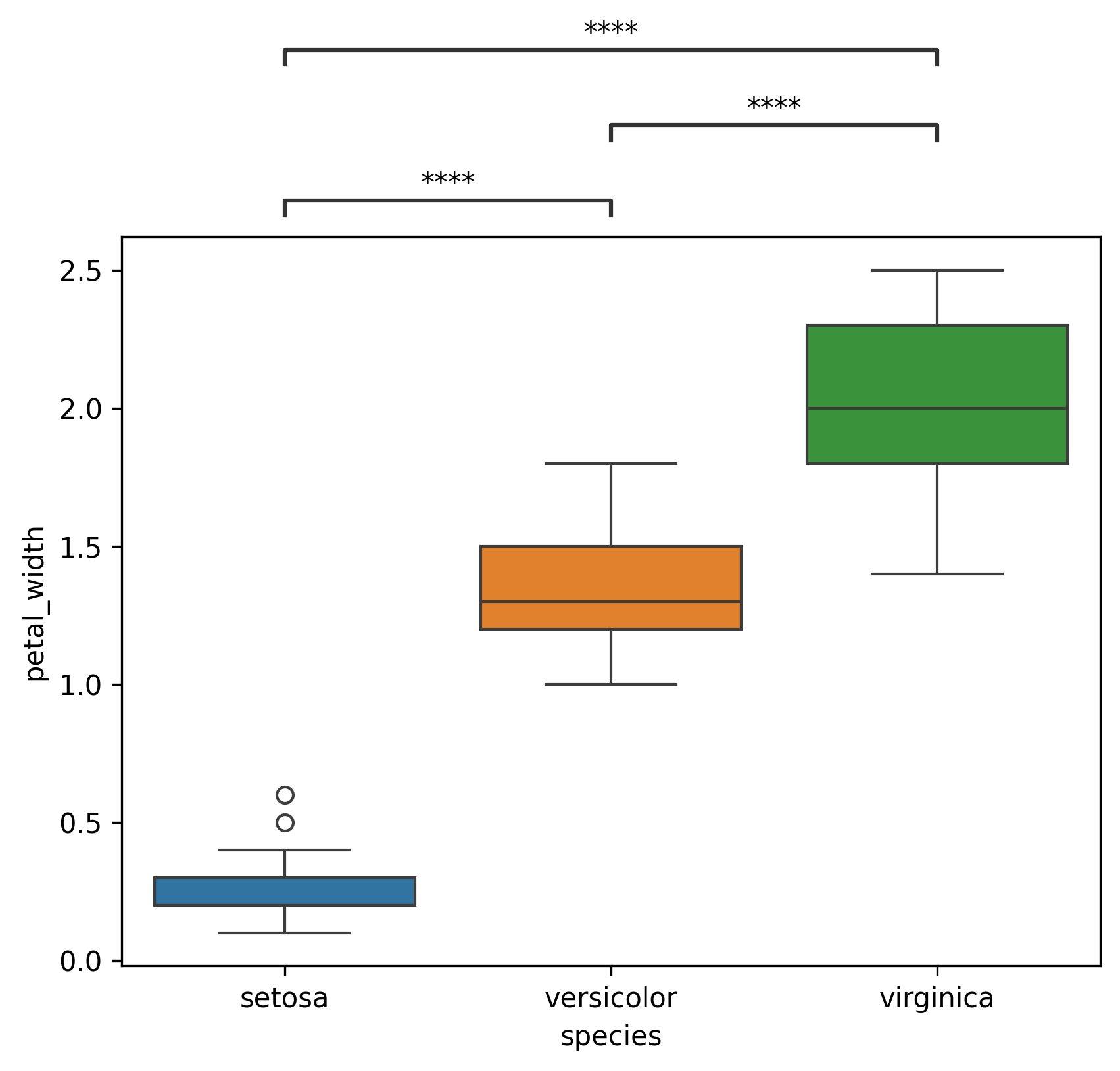
Inside (
loc='inside')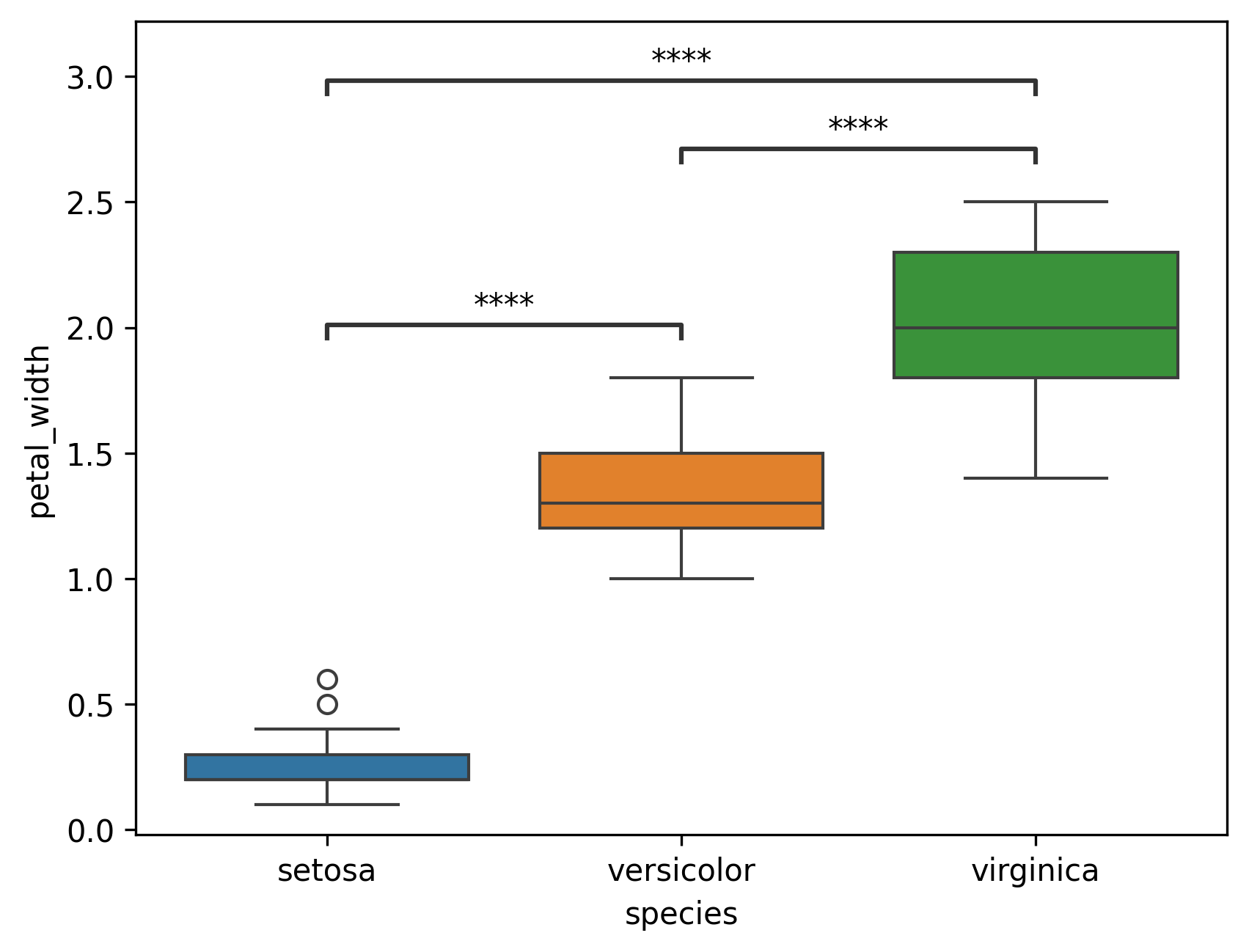
-
Alternatively, the plotting can be done using a config dictionary
Set a config dictionary
plotting = { "data": df, "x": x, "y": y, "hue": x, "hue_order": order, "order": order }Plotting
ax = sns.boxplot(**plotting)Configure annotation + multiple tests
annot.new_plot(ax, **plotting) annot.configure(comparisons_correction="Bonferroni", verbose=2)Apply test and annotate (You can daisy-chain the functions)
test_results = annot.apply_test().annotate()
Reference
- https://github.com/trevismd/statannotations